Instructors are free to include the exercises from the book in their lecture notes and course assignments. We provide here the LaTeX source code of all exercises in the book.
Chapter 1. Molecular biology and high-throughput sequencing [PDF] [LATEX]
Chapter 2. Algorithm design [PDF] [LATEX]
Chapter 3. Data structures [PDF] [LATEX]
Chapter 4. Graphs [PDF] [LATEX]
Chapter 5. Network flows [PDF] [LATEX]
Chapter 6. Alignments [PDF] [LATEX]
Chapter 7. Hidden Markov models [PDF] [LATEX]
Chapter 8. Classical indexes [PDF] [LATEX]
Chapter 9. Burrows–Wheeler indexes [PDF] [LATEX]
Chapter 10. Alignment-based genome analysis [PDF] [LATEX]
Chapter 11. Alignment-free genome analysis and comparison [PDF] [LATEX]
Chapter 12. Compression of genome collections [PDF] [LATEX]
Chapter 13. Fragment assembly [PDF] [LATEX]
Chapter 14. Haplotype analysis [PDF] [LATEX]
Chapter 15. Pangenomics [PDF] [LATEX]
Chapter 16. Transcriptomics [PDF] [LATEX]
Chapter 17. Metagenomics [PDF] [LATEX]
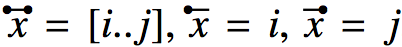
you can use the commands:
\newcommand{\fullbullet}{\begin{picture}(-1,.5)(1,-.2)\circle*{2.1}\end{picture}}
\newcommand\SP[1]{\vbox{\m@th\ialign{##\crcr
\kern.1em\fullbullet\crcr\noalign{\kern-\p@\nointerlineskip}%
$\hfil\displaystyle\overline{#1}\hfil$\crcr}}}
\newcommand\EP[1]{\vbox{\m@th\ialign{##\crcr
\hfill\fullbullet\crcr\noalign{\kern-\p@\nointerlineskip}%
$\hfil\displaystyle\overline{#1}\hfil$\crcr}}}
\newcommand\INTERVAL[1]{\vbox{\m@th\ialign{##\crcr
\kern.1em\fullbullet\hfill\fullbullet\crcr\noalign{\kern-\p@\nointerlineskip}%
$\hfil\displaystyle\overline{#1}\hfil$\crcr}}}
\makeatother